The Amino Acid Interactions (INTAA) web server is focused on investigating the energetics of amino acid interactions in the 3D structures of biomolecules. The web server's functionality is split into two services described below.
This website is free and open to all users and there is no login requirement. If you find the INTAA web server useful in your research, please cite one of the following works:
- Vymětal,J. et al. (2003) Amino Acid Interactions (INTAA) web server v2.0: a single service for computation of energetics and conservation in biomolecular 3D structures. Nucleic Acids Res., 49(W1), W15–W20, doi:10.1093/nar/gkab377
- Galgonek,J. et al. (2017) Amino Acid Interaction (INTAA) web server. Nucleic Acids Res., 45(W1), W388–W392, doi:10.1093/nar/gkx352
|
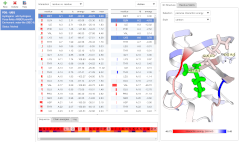
The Interaction Energy Matrix (IEM) web applicationImproved in v2.0!
This application aims to make the calculation of interaction energies between amino acid residues in proteins easily available to the public. The IEM methods are useful for determining the key residues that are important for stability and correct formation of a protein fold. It is based on the hypothesis that amino acid residues with the most stabilizing interactions contribute significantly to the folding enthalpy. Such key residues are usually structurally important and evolutionarily conserved across the homologous sequences from different organisms.
|
|
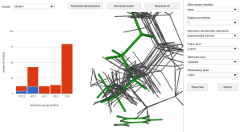
The purpose of this web server is to provide the user with the knowledge of how a specific geometrical configuration of some amino acid–DNA residue contact relates to the background of all contacts of that type. Specifically, the user can view how abundant each particular configuration is in the set of protein–DNA complexes, what is its interaction energy, and how large that interaction energy is compared to other interaction energies in that particular distribution. |

